09 Sep PhosphoPath: Visualization of Phosphosite-centric Dynamics in Temporal Molecular Networks
PhosphoPath: Cytoscape compatible visualization tool for quantitative PTM-centric proteomics data published
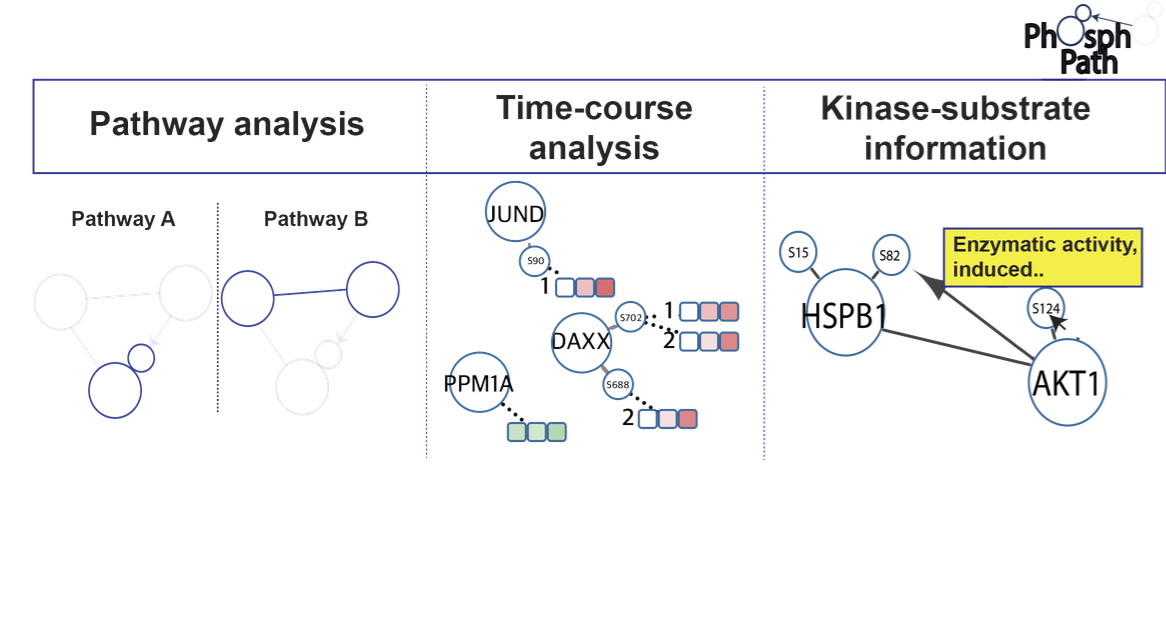
Protein phosphorylation is an essential post-translational modification (PTM) regulating many biological processes at the cellular and multicellular level. Continuous improvements in phosphoproteomics technology allow the analysis of this PTM in an expanding biological content. Yet, up till now proteome data visualization tools are still very gene centric, hampering the ability to comprehensively map and study PTM dynamics. Here we present PhosphoPath, a Cytoscape app designed for the visualization and analysis of quantitative proteome and phosphoproteome datasets. PhosphoPath brings knowledge into the biological network by importing publically available data, and enables PTM site-specific visualization of information from quantitative time series. To showcase PhosphoPath performance we use a quantitative proteomics dataset comparing patient derived melanoma cell lines grown either in conventional cell culture or xenografts.
PhosphoPath: Visualization of phosphosite-centric dynamics in temporal molecular networks
50 free e-prints of the final published article are available for interested colleagues: click here
![]()


No Comments