Welcome to the RockerBox project page
A major problem in the analysis of mass spectrometry-based proteomics data is the vast growth of data volume, caused by improvements in sequencing speed of mass spectrometers.
This growth affects analysis times and storage requirements so severely that many analysis tools are no longer able to cope with the increased file sizes.
RockerBox, addresses size problems for search results obtained from the widely used Mascot search engine by filtering the files, without altering their informative content.
Filtering is done based on manual parameters or automatically based on a number of FDR calculation methods.
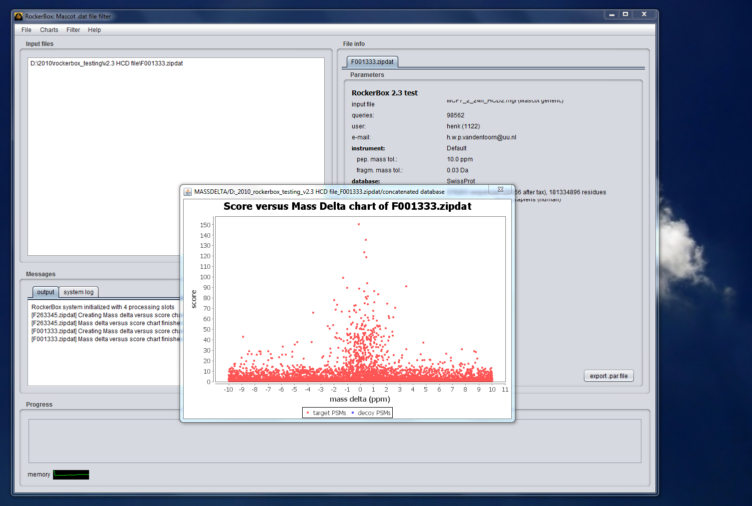
Moreover, RockerBox allows for a fast evaluation of large result files by means of a number of commonly accepted metrics which can often be viewed through charts.
(this page was previously hosted on https://trac.nbic.nl/rockerbox/wiki)

RockerBox requires Java version 8 or newer.
RockerBox has been packaged in several ways:
![]()