17 Oct Identification of Enriched PTM Crosstalk Motifs from Large-Scale Experimental Data Sets
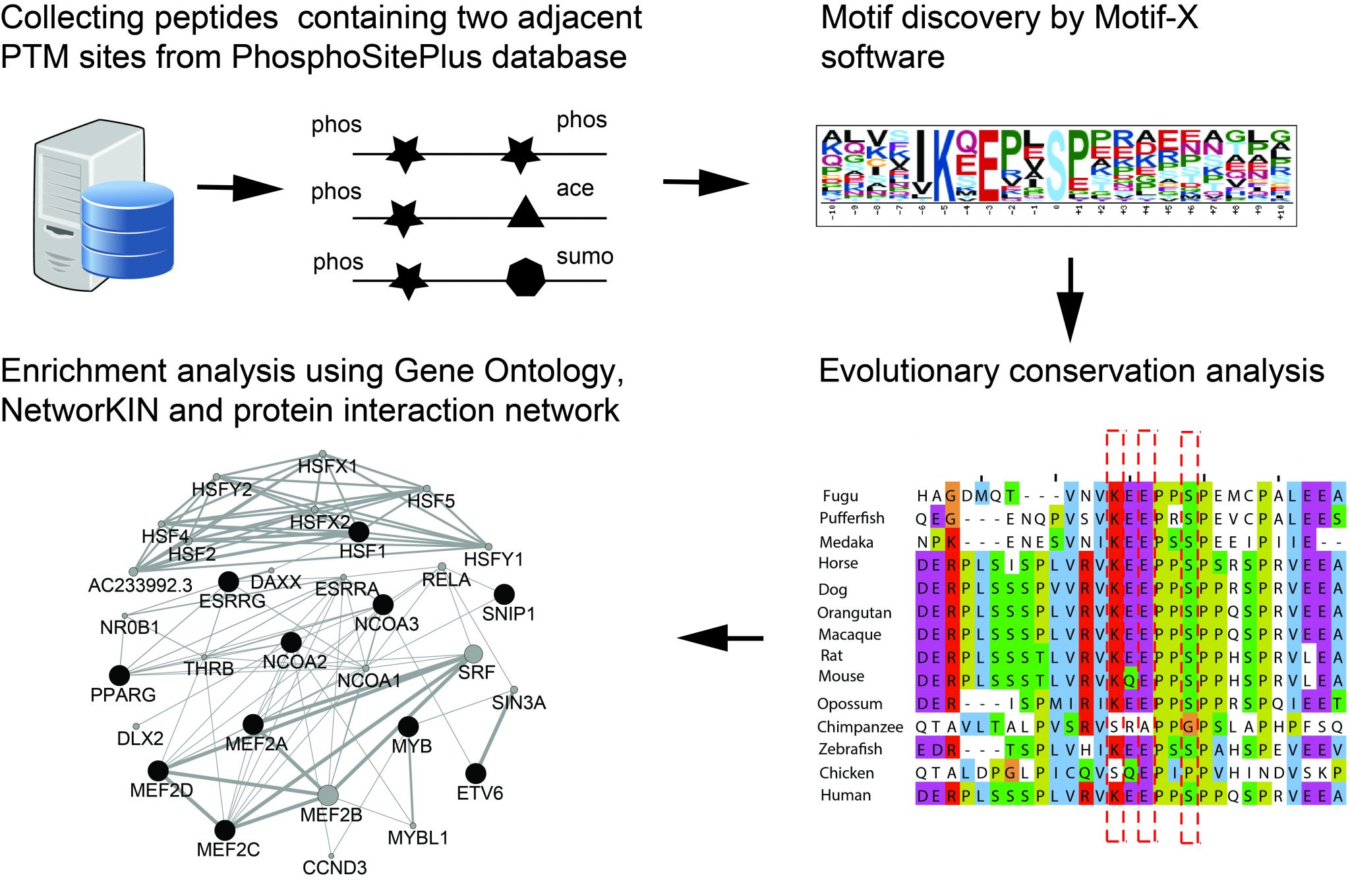
ABSTRACT: Post-translational modifications (PTMs) play an impor- tant role in the regulation of protein function. Mass spectrometry based proteomics experiments nowadays identify tens of thousands of PTMs in a single experiment. A wealth of data has therefore become publically available. Evidently the biological function of each PTM is the key question to be addressed; however, such analyses focus primarily on single PTM events. This ignores the fact that PTMs may act in concert in the regulation of protein function, a process termed PTM crosstalk. Relatively little is known on the frequency and functional relevance of crosstalk between PTM sites. In a bioinformatics approach, we extracted PTMs occurring in proximity in the protein sequence from publically available databases. These PTMs and their flanking sequences were subjected to stringent motif searches, including a scoring for evolutionary conservation. Our unprejudiced approach was able to detect a respectable set of motifs, of which about half were described previously. Among these we could add many new proteins harboring these motifs. We extracted also several novel motifs, which through their widespread appearance and high conservation may pinpoint at previously nonannotated concerted PTM actions. By employing network analyses on these proteins, we propose putative functional roles for these novel motifs with two PTM sites in close proximity.
![]()


Sorry, the comment form is closed at this time.