30 Jul Publication: Defining the background in protein network analysis; the CRAPome
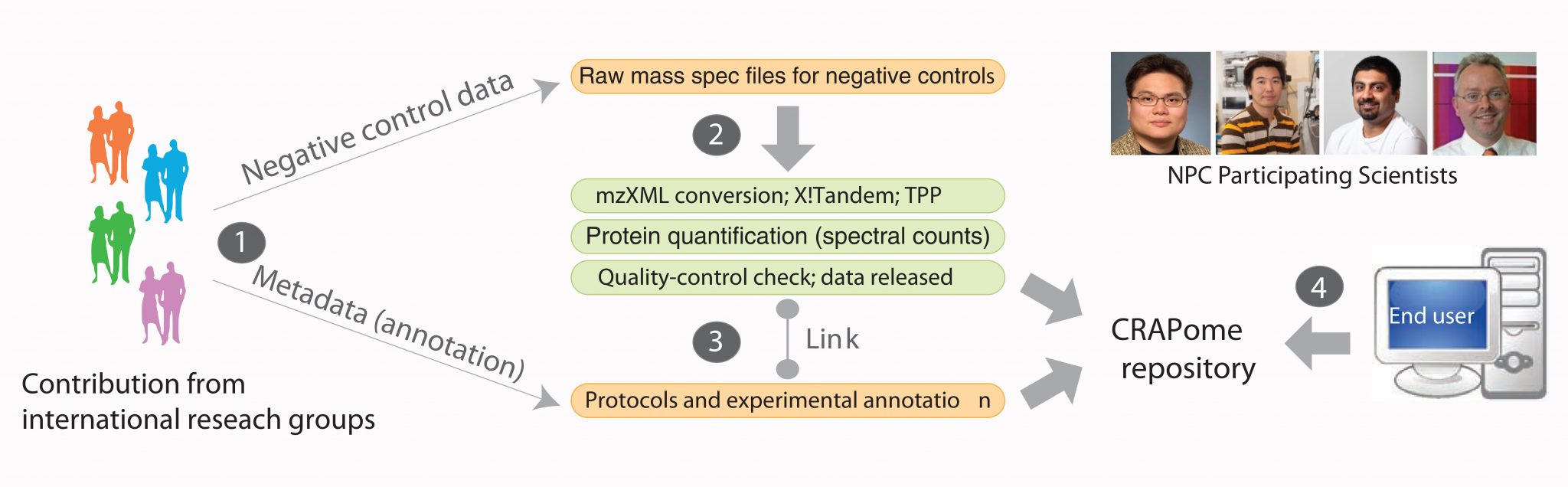
Albert Heck, Tech Yew Low, Vincent Halim and Shabaz Mohammed of the Biomolecular Mass Spectrometry and Proteomics Group (Utrecht University) joined an international effort involving many renowned proteomics researchers and cell biologists to contribute to a contaminant repository for protein affinity purifications termed the CRAPome. The CRAPome aims to create a compendium for all background contaminants derived from negative controls of AP-MS (affinity purification-MS) experiments based on various categories such as epitope tags, beads, cell lines or organisms and experimental protocols used in different labs. Although AP-MS is widely used for identifying protein-protein interactions, discriminating bona fide interactors from the background contaminants is often a daunting task. In this work, we show that, by aggregating negative controls from multiple AP-MS studies, we increase coverage and improve the characterization of background associated with a given AP-MS protocol. The current version of the CRAPome comes with online toolkits, which compute scoring functions to guide research community in decision-making. All researchers are also encouraged to contribute their negative controls to the CRAPome so as to make it an even more comprehensive resource. The CRAPome is freely available at http://www.crapome.org/.
The associated article was published in Nature Methods 7th of July 2013 (http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.2557.html).
![]()



Sorry, the comment form is closed at this time.