12 Jun Publication in Cell Reports
Last year two first drafts of the human proteome were made available [1,2]. These drafts provide an overview of all proteins detected in different cells and tissues of our body. As proteins are the prime molecular entities responsible for all biological processes these drafts tell us which “players” are present. A next step in the analysis of the proteome is to observe how proteins work and communicate with each other. They most often do this by the transfer of a chemical phosphate moiety from one to the other protein, which may lead to activation or deactivation of the protein modified.
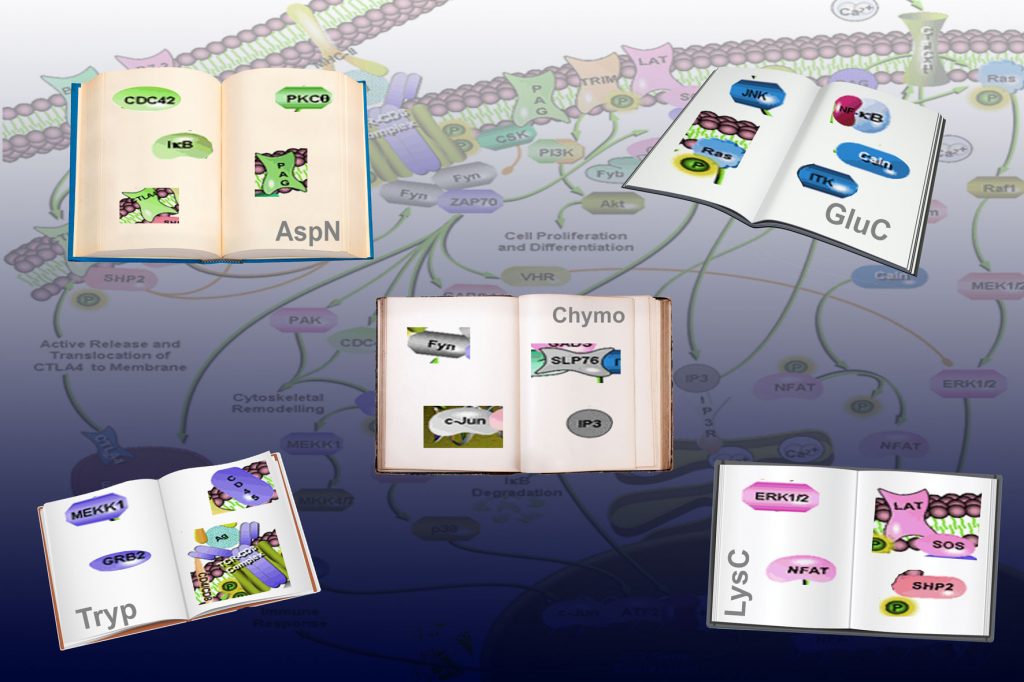
Proteomics researchers have been aiming at unraveling this communication language, termed the phosphoproteome. But as Piero Giansanti et al. [3] show in the latest issue of Cell Reports, these researchers have been using a single dictionary (i.e. workflow) for that, which can only help to translate a specific part of the communication. By using 5 different dictionaries (5 different workflows) Giansanti et al. are able to provide a much broader view into cellular communication, and show indeed that in the public repositories there is a considerable bias in the phosphoproteome. Each dictionary was able to translate about 70% new parts of communication, whereas only 7% of what the cells were communicating could be translated by all 5 different workflows. More technically, the 5 “dictionaries” are presented by 5 different proteases that were used to cleave the proteins into for analysis amendable peptide sequence reads. Using this multiple protease they clearly showed that optimal detection of each protein individual phosphorylation event is linked to a preferred protease.
In the proteolysis step the greatest loss occurs as many of the generated small peptides, get lost in translation. Giansanti et al. repair this problem and present an augmented human phosphopeptide atlas of 37,771 unique phosphopeptides, each representing a “word of communication” in the cell.
1. Kim, M. S., Pinto, S. M., …. & Pandey, A. (2014) A draft map of the human proteome, Nature. 509, 575-81.
2. Wilhelm, M., Schlegl, J., ….. & Kuster, B. (2014) Mass-spectrometry-based draft of the human proteome, Nature. 509, 582-7.
3. Giansanti, P, Aye, T.T., van den Toorn, H., Peng, M., van Breukelen, B. & Heck A.J.R (2015) An augmented multiple protease based human phosphopeptide atlas, Cell Reports
You can listen to the radio interview with Albert Heck about this research project here
![]()


Sorry, the comment form is closed at this time.